- Visibility 421 Views
- Downloads 38 Downloads
- DOI 10.18231/j.jdpo.2024.042
-
CrossMark
- Citation
Deep learning models for tumor detection and segmentation in medical image analysis: A comprehensive review of ResNet, U-Net, DETR, and inception variants
- Author Details:
-
Naseebia Khan *
-
Abhinaba Das
Introduction
Medical images are vital for diagnosing and treating diseases.[1] However, the traditional manual reading of images can be slow and prone to errors. With the advancement of technology, deep learning has become a valuable tool in various fields, including medicine. Deep learning improves the efficiency and accuracy of image-based diagnosis, making it an important part of modern medical practice. Deep learning models offer several advantages in medical imaging. They can improve the accuracy of diagnosis by identifying patterns that may be missed by human eyes. Additionally, these models work efficiently and can analyze medical images much faster than humans, leading to quicker diagnoses and treatment decisions. Moreover, the consistency of deep learning models ensures reliable and unbiased results, reducing the risk of mistakes caused by human factors. Overall, deep learning plays a vital role in enhancing medical image analysis and patient care.
Radiologists encounter several challenges in traditional image analysis, including the time-consuming and labor-intensive nature of manual image review. Each image requires scrutiny, leading to potential delays in diagnosis and treatment decisions, especially in settings with a high volume of imaging data. Additionally, human interpretation can be subjective, resulting in inter-observer variability, where different radiologists may provide varying diagnoses for the same image. This variability poses a risk to the consistency and reproducibility of image analysis. Moreover, as the field of medical imaging advances, traditional methods may struggle to handle the increasing complexity and diversity of patterns observed in medical images, particularly in rare or novel cases.
Deep learning models
The recent application of Convolutional Neural Networks(CNNs), a field of deep learning (DL), has brought about remarkable advancements in radiology, as highlighted by Hinton in JAMA(2018) and LeCun et al. in Nature (2015).[2] These DL-based models have demonstrated significant promise in the detection of nodules/masses on chest radiographs.[3], [4], [5], [6], [7] In clinical practice, distinguishing between benign and malignant nodules and detecting them accurately presents challenges for radiologists. The resemblance of normal anatomical structures to nodules can lead to misinterpretation, necessitating careful attention to nodule characteristics. Interestingly, the issues often stem from the complexities of the cases rather than the radiologists' skill, making misdiagnosis possible even among experienced practitioners. [8], [9]
Convolutional Neural Networks (CNNs), as introduced by LeCun et al. in 1989[10] and popularized by Krizhevsky et al. in 2012, have brought transformative advancements to medical imaging.[10], [11] These networks have revolutionized the field by harnessing their capacity to autonomously learn intricate data representations. The renaissance of CNNs has yielded remarkable progress across diverse medical imaging modalities, including Radiography,[12] Endoscopy,[13] Computed Tomography(CT).[14], [15] Mammography Images(MG).[16] Ultrasound Images, [17] Magnetic Resonance Imaging(MRI).[18], [19] and Positron Emission Tomography(PET).[20] These advancements underscore the versatility and potential of CNNs to elevate image analysis across a spectrum of medical imaging domains ([Figure 1]).
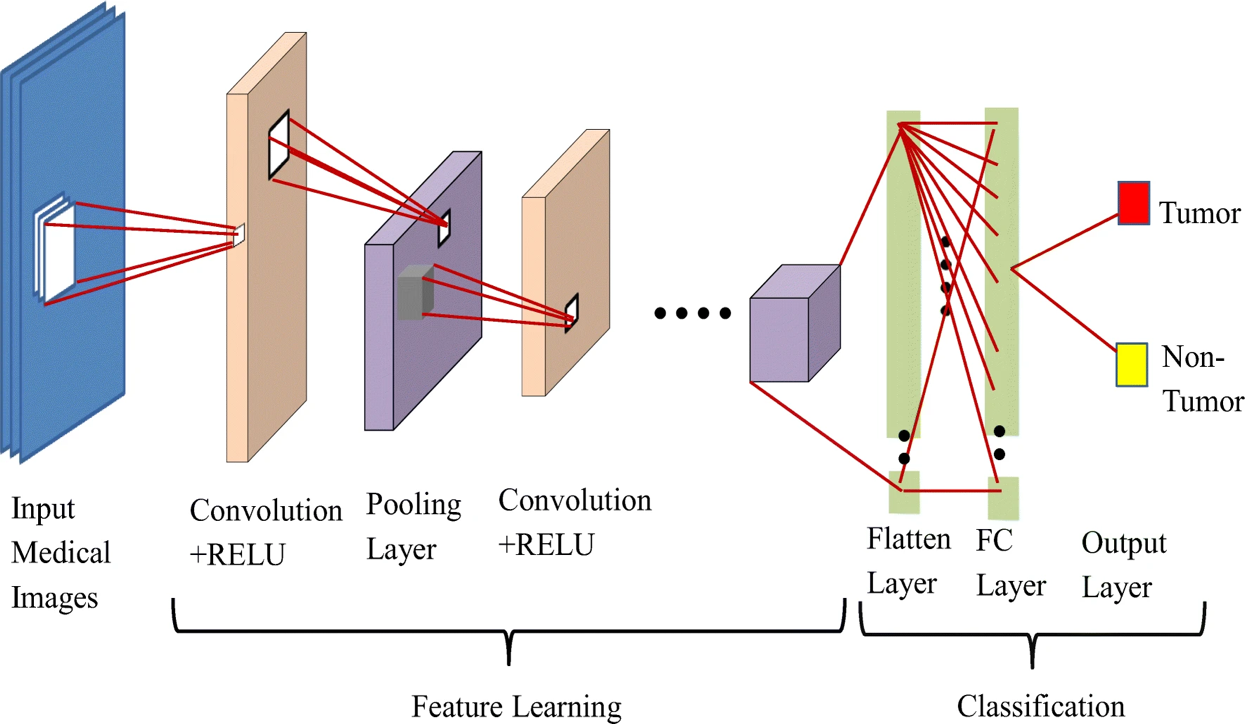
Deep learning models present compelling solutions to these challenges in medical image analysis. By automating image interpretation, these models can expedite the process, allowing radiologists to focus on more complex cases and expedite patient care. The consistency and reproducibility of deep learning results reduce inter-observer variability, leading to more reliable diagnoses. Furthermore, deep learning models are scalable and can handle large datasets efficiently, making them suitable for real-world clinical applications. The ability of these models to learn intricate patterns from extensive data enables them to identify complex features that might be overlooked by human observers. As a result, deep learning models not only enhance the accuracy of image analysis but also contribute to improved patient outcomes and better healthcare delivery.
Deep learning models have emerged as powerful allies to radiologists in overcoming the challenges of traditional image analysis. Their automation, efficiency, consistency, scalability, and ability to learn complex patterns make them indispensable tools for modern medical imaging. By embracing these advanced techniques, the medical community can revolutionize image-based diagnosis and treatment, ultimately benefiting patients worldwide. Nevertheless, ongoing research, validation, and collaboration between deep learning experts and medical practitioners remain crucial to ensure the responsible integration and optimal utilization of these models in clinical settings.
Overview of tumor detection and segmentation
In the realm of tumor detection and segmentation using deep learning, several state-of-the-art models have emerged as prominent players. U-Net, a purpose-built architecture for biomedical image segmentation, stands out for its encoder-decoder structure with skip connections that adeptly integrates fine-grained features during upsampling, enabling accurate tumor boundary delineation. ResNet, renowned for introducing residual connections, offers a robust feature learning capability that has been harnessed for tumor classification and localization tasks. Meanwhile, DETR, a transformer-based model initially designed for object detection, has made strides in medical image analysis, showcasing its prowess in instance segmentation tasks such as tumor localization. Inception variants, notably Inception-v3 and Inception-ResNet, capitalize on multi-scale convolutional filters to excel in tumor detection and classification across various resolutions.
Each deep learning model has unique strengths that make it suitable for specific tumor detection and segmentation tasks. The choice of the model depends on the specific requirements of the application, the available data, and the desired level of accuracy and interpretability. Furthermore, ensembling or combining multiple models could further enhance the performance and robustness of tumor detection and segmentation systems. As deep learning continues to evolve, further research and innovations are expected to yield even more effective models for medical imaging tasks in the future.
The integration of state-of-the-art deep learning models for tumor detection and segmentation holds significant promise and potential impact in medical imaging. These models, such as U-Net, ResNet, DETR and Inception variants, can revolutionize clinical practices by enhancing the accuracy, efficiency, and consistency of tumor analysis across diverse medical imaging modalities including MRI, CT, and histopathological slides. Their integration improves patient care through early detection, personalized treatment strategies, and effective monitoring of treatment progress. The scope extends to a wide range of tumor types, providing radiologists and clinicians with valuable tools for comprehensive and reliable analysis. Moreover, the exploration of ensembling multiple models can further expand the scope by harnessing synergistic benefits and encompassing a broader spectrum of tumor detection and segmentation challenges.
However, it is essential to address certain limitations inherent in the review process. The rapid pace of advancements in deep learning may result in the exclusion of newer models not covered in the review. This limitation underscores the evolving nature of the field and the continuous emergence of novel techniques. Additionally, the availability of high-quality, diverse, and well-annotated medical imaging datasets is crucial for a comprehensive evaluation of these models' effectiveness. The absence of certain datasets may impact the robustness and generalizability of the review's findings. Moreover, variations in evaluation metrics and experimental setups across different studies can introduce challenges in direct model comparison. Acknowledging and addressing these limitations ensures a well-rounded understanding of the scope and potential constraints of integrating deep learning models for tumor detection and segmentation in medical imaging.
Deep learning fundamentals
Deep learning is a powerful branch of machine learning centered on training neural networks to identify patterns in data. Neural networks consist of interconnected nodes, with training involving weight adjustments to minimize prediction errors. Convolutional Neural Networks(CNNs), tailored for image analysis, use convolutional and pooling layers to extract features and activation functions to capture complex relationships. CNNs are pivotal in image tasks like classification, object detection, and segmentation.
Transformer-based models, initially designed for sequential data like language, have found adaptation in medical image analysis. These models excel at capturing long-range dependencies and relationships. Inspired by the self-attention mechanism, transformers aid in holistic understanding, crucial for tasks such as tumor detection and organ segmentation in medical images. The fusion of deep learning's capabilities and transformer models' adaptability showcases their immense potential in revolutionizing medical image analysis.
ResNet and variants
Overview of ResNet architecture
The Residual Network(ResNet) architecture, introduced by Kaiming He et al. [21] is a pivotal advancement in deep learning due to its ability to train very deep neural networks without encountering the vanishing gradient problem. ResNet incorporates residual blocks that consist of identity shortcuts, allowing the network to learn residual mappings. This enables the network to focus on learning the difference between the input and the desired output, making training more efficient. Different versions of ResNet, such as ResNet-18, ResNet-34, ResNet-50, and ResNet-101, vary in their depth, with deeper versions having more layers. Deeper networks theoretically can capture finer features and representations. However, it has been observed that overly deep networks may suffer from degradation in performance due to optimization difficulties. ResNet-50 is a popular choice as it strikes a balance between depth and performance([Figure 2]).
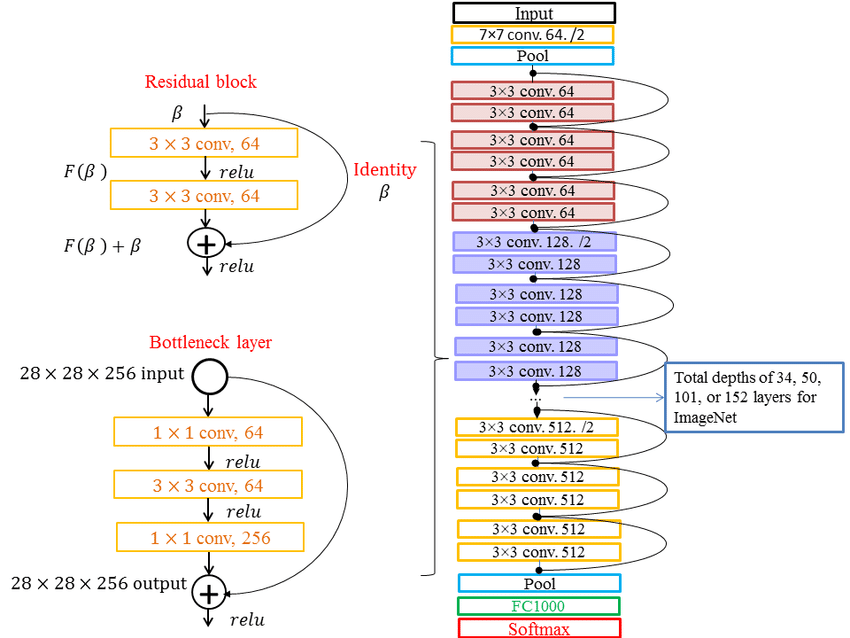
Applications of ResNet in tumor detection and segmentation
ResNet has shown remarkable promise in tumor detection and segmentation across various imaging modalities and tumor types. In recent years, the spotlight in deep learning research has illuminated the significance of residual neural networks(ResNet) and their optimization, with a strong presence in the field of medical images. ResNet's applications have been particularly impactful in clinical realms such as diagnosis, staging, metastatic evaluation, therapy planning, and target selection for severe conditions including tumors, cardiovascular and cerebrovascular disorders, and nervous system ailments. Noteworthy contributions to this landscape include Li et al. (2018),[22] who introduced a dense convolutional neural network-based classification method for Alzheimer's Disease(AD) classification by leveraging MRI brain images to learn diverse local characteristics. Additionally, Liu et al. [22] developed a multi-scale residual neural network that adeptly collects multi-scale information from images and applies residual learning, showcasing the network's ability to capture intricate multi-scale features.
Further extending the utilization of ResNet, Murad et al. [23] proposed a 3D deep residual neural network, tailored for brain anatomy analysis using 3D images, and harnessed it to build a brain age prediction model. Nibali et al. [24] harnessed ResNet for categorizing benign and malignant lung nodules, investigating the impact of transfer learning and various network depths on the accuracy of malignant tumor classification. Collectively, these applications highlight the diverse and powerful applications of ResNet in medical image analysis, addressing challenges across various clinical domains.
Multi-scale residual neural network(MSResNet) is used to extract multi-scale information from convolution kernel images of varying sizes and conducted residual learning on the neural network. This method enabled multi-scale feature learning, resulting in improved classification compared to traditional deep convolutional networks. Highlighting the profound influence of optimal learning algorithms, the success of a residual neural network, as noted by Maier et al. [25] has significantly contributed to advancements in medical image analysis.
A distinctive contribution to medical image analysis is ResNet-22, designed exclusively for breast cancer screening categorization, their model extends the common ResNet architecture with a depth-to-width ratio optimized for analyzing high-resolution medical images, yielding promising experimental outcomes. Further showcasing ResNet's efficacy, Karthik et al.[26] demonstrated its application in identifying COVID-19 in chest X-ray images. Moreover, Lu et al. [27] leveraged ResNet and UNet++ to propose the WBC-Net deep learning network, incorporating a hybrid skip route using dense convolutional blocks for multi-scale data aggregation and a context-aware feature encoder employing residual blocks for multi-scale feature extraction. This innovative approach led to enhanced accuracy in white blood cell image segmentation. Additionally, Nazir et al. [28] introduced the OFF-eNET architecture, amalgamating residual mapping and Inception modules for automatic intracranial vessel segmentation. The architecture achieved a richer visual representation while improving computational efficiency.
In lung cancer detection, studies like "Deep Learning-Based Detection System for Multiclass Lesions on Chest Radiographs" have demonstrated ResNet's ability to outperform human radiologists in certain scenarios. [4] In breast cancer analysis, "Deep Residual Learning for Mammographic Mass Classification" showcased ResNet's superiority in mammography mass classification. [29] ResNet variants have also been applied to brain tumor segmentation, with the "3D Deep Residual Networks for Glioma Segmentation in MRI" achieving impressive results.[30] These studies highlight ResNet's potential to significantly improve tumor detection and segmentation performance, outperforming traditional methods and rivaling other deep learning approaches.
Variants and modifications of ResNet for tumor analysis
Various modifications and enhancements have been made to ResNet for tumor analysis tasks. Attention mechanisms, like in "Attention Residual Learning for Skin Lesion Classification", enhance ResNet's capability to focus on relevant features, leading to improved classification accuracy. Skip connections at different depths, as demonstrated in "Improving Automated Melanoma Recognition using Deep Learning Residual Networks", aid in gradient propagation and training stability. Transfer learning, drawing from pre-trained models on large datasets, has shown substantial benefits in tasks like brain tumor segmentation("Brain Tumor Segmentation Using Residual U-Net" by Akkus et al. [19]), enabling effective feature extraction and generalization.
These modifications have shown a positive impact on model performance, enhancing accuracy and robustness in tumor detection and segmentation tasks. Additionally, they contribute to improved training efficiency by mitigating issues like vanishing gradients and convergence challenges. Overall, ResNet and its adaptations continue to shape the landscape of medical imaging by bolstering the accuracy and efficiency of tumor analysis tasks.
U-Net and variants
U-Net architecture
The U-Net architecture, introduced by Ronneberger et al. [31] in 2015, is a deeply influential convolutional neural network(CNN) design tailored for semantic segmentation tasks, particularly in the medical imaging domain. U-Net's distinctive architecture comprises an encoding path, responsible for feature extraction through convolutional and pooling layers, and a decoding path, which employs upsampling and transposed convolutions for feature map expansion. The encoding path captures context and spatial information, while the decoding path recovers spatial resolution, enabling precise segmentation. Key to U-Net's success is skip connections that bridge corresponding layers in the encoding and decoding paths, facilitating the fusion of high-level semantics and fine-grained spatial details. This enables U-Net to effectively capture high-resolution features while preserving global context ([Figure 3]).

Segmentation methods offer enhanced insight compared to detection techniques. In clinical settings, achieving pixel-level lesion size classification improves diagnostic accuracy and facilitates accurate tracking of morphological changes. This approach, as emphasized by Schwartz et al. [32] allows for a comprehensive assessment of lesion characteristics, encompassing dimensions and area, crucial for evaluating treatment effects.
U-Net for limited labeled data
U-Net's design is particularly advantageous for semantic segmentation tasks with limited labeled data. Its architecture inherently enables the network to learn from sparse annotations, as skip connections enable gradient flow to both high and low-resolution features. This property is crucial when labeled data is scarce, as U-Net learns to leverage limited annotations effectively. Additionally, U-Net's contracting and expanding pathways aid in learning hierarchical representations, which is advantageous when a small dataset cannot provide a diverse range of examples.
Applications of U-Net in tumor detection and segmentation
U-Net's applications in tumor detection and segmentation have yielded significant advancements. For instance, in breast cancer analysis, "A Novel U-Net Based Fully Convolutional Network for Breast Density Segmentation" employed U-Net to segment breast density. In glioma segmentation, "Glioma Segmentation in Brain MRI Images Using U-Net" showcased its effectiveness. U-Net has been applied to lung nodule segmentation as well, as seen in "Automatic Pulmonary Nodule Detection via 3D U-Net".[33] Similarly, Liu et al.[34] also propose a hybrid architecture consisting of transformer layers in the decoder part of 3D UNet to accurately segment tumors from volumetric breast data.[31] U-Net's segmentation capabilities in diverse tumor types and imaging modalities have been essential for improving diagnostic accuracy.
Adaptations and enhancements of U-Net
U-Net variants have emerged to address specific tumor segmentation challenges. Multi-scale feature integration, as in "MultiResUNet: Rethinking the U-Net architecture for multimodal biomedical image segmentation", improves performance by integrating features from different scales. Attention mechanisms, as in "Attention U-Net: Learning Where to Look for the Pancreas", enhance the model's focus on relevant regions. Data augmentation strategies like "Augmenting Data for Liver Lesion Classification using U-Net-based Autoencoder" mitigate data scarcity. In addition, 3D U-Net is an easy adaptation of U-Net for 3D image segmentation. [31] Combining the 3D U-Net network with the residual network's residual blocks create a new 3D residual U-Net network. These adaptations showcase U-Net's versatility and its ability to be tailored to specific challenges in tumor segmentation.
DETR(Detection Transformer)
DETR architecture and object detection
The DETR(DEtection TRansformers) architecture, introduced by Carion et al. in 2020,[35] revolutionizes object detection by replacing the conventional anchor-based methods with a transformer-based framework. Traditional object detection approaches relied on predefined anchor boxes to predict object locations, which posed challenges in handling scale, aspect ratio variations, and anchor design. DETR addresses these limitations by casting object detection as a set prediction problem. It employs a transformer encoder to capture global context and a transformer decoder to predict object instances directly without anchors. Position embeddings facilitate precise localization. By introducing bipartite matching and set-based loss functions, DETR aligns predicted instances with ground-truth objects, enabling end-to-end training for detection tasks([Figure 4]).
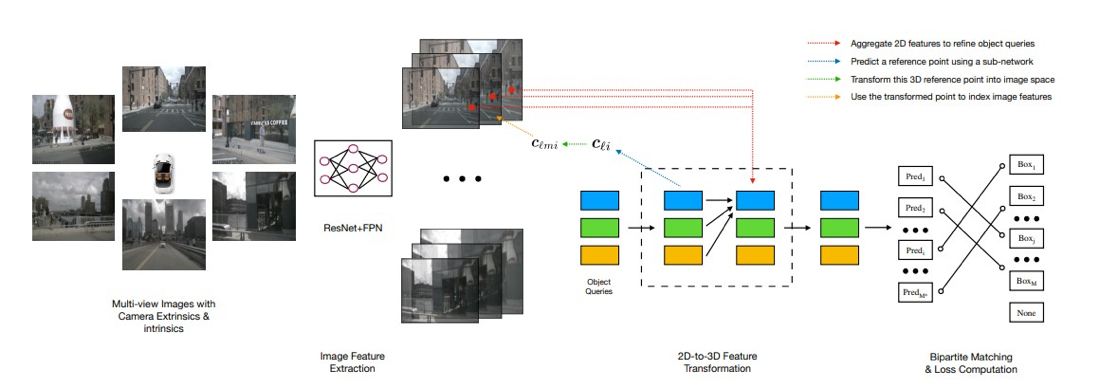
Applications of DETR in Medical Image Analysis: DETR has found promising applications in medical image analysis, particularly in tumor detection and localization.In the realm of medical detection, leveraging deep learning methods to enable machines for autonomous feature learning in images and the identification of anomalous regions holds significant importance. However, the challenge persists in effectively detecting small lesions within medical images due to the diminutive size of the objects, necessitating the machine's adeptness in filtering out background information and precisely pinpointing these minute anomalies. Recent advancements in research have introduced pivotal solutions. Carion et al.[35] proposed DETR, combining CNN for feature extraction and Transformer for encoding and decoding to predict bounding boxes. Vision Transformer(ViT) is introduced employing self-attention for image feature extraction by dividing images into patches. Liu et al. [34] extended ViT to Swin Transformer, enhancing image recognition and object detection by optimizing the self-attention mechanism through a window-based approach.
In the context of instance segmentation, DETR's ability to handle varying object sizes and shapes without anchor priors is advantageous. Studies such as "DETR for Automated Lung Nodule Detection in CT Scans" have demonstrated DETR's effectiveness in detecting lung nodules. Its instance segmentation capabilities, as seen in "Tumor Segmentation in Breast Ultrasound Images Using DETR", showcase its potential in segmenting tumors with varying shapes and sizes. DETR's holistic approach allows it to excel in medical image analysis tasks where objects exhibit diverse appearances.
Advantages and limitations of using DETR
DETR's elimination of anchor-based design simplifies model training and design, making it more intuitive. Its inherent ability to handle varying object sizes and layouts contributes to its versatility. However, DETR may require larger amounts of data to achieve optimal performance due to its reliance on end-to-end training. Additionally, while DETR is highly capable, its processing speed can be slower than traditional methods, making it less suitable for real-time applications.
Adaptations and extensions of DETR for tumor detection and segmentation
DETR's adaptability extends to tumor detection and segmentation challenges. Modified loss functions, as demonstrated in "Improved DETR for Liver Lesion Detection and Segmentation", enhance its performance for specific tasks. Incorporating domain-specific priors or pretraining on medical images can further fine-tune DETR for medical applications. Such adaptations can improve the model's capability to handle medical image intricacies and lead to more accurate tumor detection and segmentation.
Inception and variants
The Inception architecture([Figure 5]), introduced by Szegedy et al. [36] is known for its efficient multi-scale feature extraction using parallel convolutional layers of various kernel sizes. The architecture aims to capture features at different scales by processing input through multiple convolutions of different receptive fields simultaneously. The hallmark of the Inception architecture is the Inception module, which incorporates convolutional filters of varying sizes(1x1, 3x3, 5x5) concatenated together, alongside max-pooling operations. This parallel processing across different kernel sizes enables the network to learn intricate details and capture both local and global patterns within the data.
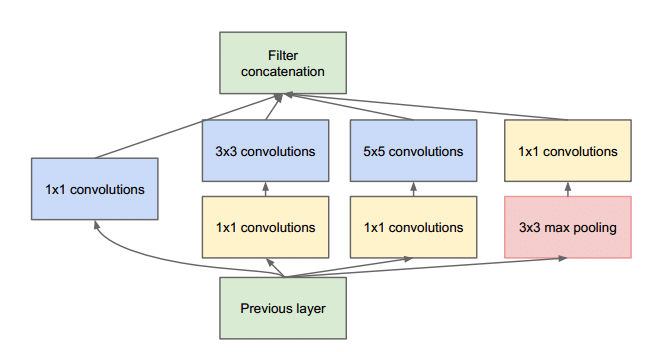
Inception module and computational complexity
The Inception module serves as the building block of the Inception architecture. Its unique design allows for feature extraction across different scales while reducing computational complexity. This is achieved through 1x1 convolutions, which act as bottleneck layers, reducing the number of input channels before applying more computationally intensive larger convolutions. By reducing the dimensionality early in the process, the Inception module helps mitigate the computational burden associated with processing multiple convolutions in parallel. This reduction in computational complexity is critical in enabling deep networks like Inception to scale effectively while maintaining high performance.
Applications of Inception Models in Tumor Analysis: Inception models have been effectively applied to tumor detection and segmentation across various imaging modalities and tumor types. For instance, "A Hybrid Approach for Brain Tumor Detection using Inception-v4 Network" employed Inception-v4 for brain tumor detection, showcasing its capability to discern tumor regions from medical images. In "Breast Tumor Segmentation and Classification using Inception-ResNet-v2", Inception-ResNet-v2 was utilized for breast tumor segmentation, highlighting its potential in segmenting intricate tumor boundaries. These applications underscore the Inception architecture's versatility in improving accuracy and efficiency in tumor analysis tasks.
Performance of inception models
Inception models have demonstrated notable advancements in tumor analysis compared to other deep learning architectures and traditional methods. Studies have shown that Inception models, with their multi-scale feature extraction capabilities, outperform traditional methods in detecting and segmenting tumors across diverse medical imaging modalities. When compared to conventional CNN architectures, Inception models exhibit enhanced efficiency in capturing fine-grained details and contextually significant features, leading to improved accuracy in tumor analysis tasks.
Variants and customizations of inception models for medical imaging
To optimize Inception models for medical imaging, researchers have introduced adaptations that augment their performance. Attention mechanisms, like the "Attention Inception Network for Brain Tumor Segmentation”, have been integrated to enhance the focus on tumor regions, improving the models' ability to identify critical areas. Specialized Inception modules, designed to cater specifically to medical imaging characteristics, have been proposed in studies like "Medical Image Segmentation using Specialized Inception Modules", emphasizing the tailored nature of these models. Moreover, fine-tuning hyperparameters, such as kernel sizes and depth, has been explored in "Optimizing Inception Models for Lung Tumor Detection", demonstrating the significance of customization for medical tasks. [37]
Benefits of inception variants in tumor analysis
Empirical studies have highlighted the benefits of customized Inception variants in tumor analysis. Incorporating attention mechanisms has led to improved focus on tumor regions, effectively reducing false positives and enhancing segmentation accuracy. Specialized Inception modules have proven to be particularly effective in handling medical image intricacies, contributing to more accurate tumor detection and segmentation. Optimized hyperparameters have further fine-tuned Inception models to yield superior performance in detecting tumors within specific medical contexts. These tailored adaptations underscore the potential of Inception variants to significantly enhance tumor analysis outcomes.
Comparative analysis
Commonly Used Evaluation Metrics for Tumor Detection and Segmentation: Several evaluation metrics are commonly used to assess the performance of tumor detection and segmentation models:
Dice Coefficient(DSC): The Dice coefficient measures the overlap between the predicted and ground truth segmentation masks. It ranges from 0 to 1, where 1 indicates perfect overlap. It is calculated as DSC=∣A∣+∣B∣2×∣A∩B∣, where A is the predicted mask and B is the ground truth mask.
Sensitivity(True Positive Rate): Sensitivity measures the model's ability to correctly detect true positive cases. It is calculated as TP/TP+FNTP, where TP is the number of true positives and FN is the number of false negatives.
Specificity(True Negative Rate): Specificity measures the model's ability to correctly identify true negative cases. It is calculated as TN/TN+FPTN, where TN is the number of true negatives and FP is the number of false positives.
Intersection over Union(IoU): IoU measures the overlap between the predicted and ground truth regions, normalized by the total area. It is calculated as IoU=∣A∪B∣∣A∩B∣, where A is the predicted region and B is the ground truth region.
Comparative analysis of deep learning models for tumor analysis
ResNet, U-Net, DETR, and Inception variants offer diverse strengths and considerations for tumor analysis tasks. ResNet's ability to learn deep representations makes it suitable for capturing complex features, while U-Net's encoding-decoding structure excels in segmenting small structures. DETR's transformer-based framework handles object detection and instance segmentation effectively. Inception variants with multi-scale feature extraction are versatile.
In terms of performance metrics, ResNet and Inception variants, with their feature extraction capabilities, achieve high DSC and IoU scores. U-Net's specialization in segmentation yields excellent results for small structures. DETR's object detection accuracy is prominent. Regarding model complexity, ResNet and Inception models tend to be deeper, demanding substantial computational resources. U-Net's compact architecture simplifies training. DETR's transformer design adds complexity but enhances instance recognition.
Training time varies; U-Net's simplicity facilitates quick training, while ResNet and Inception's depth increases training duration. DETR's transformer design might require longer training, yet its end-to-end approach is efficient.
Suitability varies based on tumor types and modalities. U-Net's segmentation power suits well for smaller, intricate structures like nodules. ResNet's and Inception's feature extraction prowess benefits tumor identification. DETR excels in object detection, well-suited for cases with diverse tumor shapes.
ResNet
ResNet, with its deep architecture, excels in tumor detection and segmentation tasks by learning complex features and capturing intricate patterns. It achieves high accuracy, particularly in cases with nuanced features. However, its deeper structure contributes to higher model complexity, demanding substantial computational resources for training and deployment. Additionally, the depth of ResNet might lead to overfitting, especially when training data is limited.
U-Net
U-Net's distinctive encoding-decoding structure makes it exceptionally effective in segmenting small structures like tumors with precision. Its compact design offers lower computational complexity, making it well-suited for scenarios with limited resources. On the flip side, U-Net's architecture might struggle with capturing global context, potentially missing larger context in tumor images. Moreover, while U-Net excels in segmentation, its primary focus isn't object detection, which could be a limitation for tasks requiring both.
DETR
DETR's transformer-based framework is particularly advantageous for object detection tasks, making it suitable for identifying tumors of varying shapes and sizes. Its end-to-end learning framework simplifies training and eliminates challenges associated with anchor-based designs. However, the transformer architecture increases model complexity, potentially leading to longer training times. Additionally, DETR might face difficulties in detecting fine details within tumors due to its primary focus on object-level detection.
Inception models
Inception models' unique parallel convolution approach captures multi-scale features effectively, enhancing their ability to discern diverse tumor patterns. They strike a balance between accuracy and efficiency, making them suitable for a wide range of tasks. Nevertheless, the incorporation of multiple parallel paths increases model complexity, necessitating careful resource allocation. The trade-off between multiple paths might also lead to shallower networks compared to ResNet, potentially affecting accuracy in certain scenarios.
Trade-offs and considerations
Selecting the appropriate model entails trade-offs. While higher accuracy often comes with increased model complexity and longer training times, models like U-Net emphasize efficiency and simplicity. There's a balance between accuracy and the global context captured, as seen in U-Net and ResNet. The choice ultimately depends on the specific task's requirements, dataset size, and available resources, while considering the potential trade-offs between accuracy, efficiency, and model complexity.
Datasets and benchmarking
The listed datasets, among others, serve as valuable resources for the development and evaluation of deep learning models in medical image analysis. They contribute to advancing the field of computer-aided diagnosis and treatment planning for various medical conditions.
BraTS(brain tumor segmentation)
The BraTS dataset, which stands for Brain Tumor Segmentation, is a widely used dataset for training and evaluating deep learning models in the field of neuroimaging. It provides MRI scans of brain tumors along with ground truth segmentation masks for tumor regions. The dataset includes different tumor types, such as gliomas and meningiomas, and contains multiple modalities like T1-weighted, T2-weighted, and FLAIR images. Researchers utilize this dataset to develop models for accurate tumor segmentation and classification, aiding in diagnosis and treatment planning for brain tumor patients.
LIDC-IDRI(lung nodule detection)
The LIDC-IDRI dataset, or Lung Image Database Consortium and Image Database Resource Initiative, is commonly employed for deep learning tasks related to lung nodule detection and classification in computed tomography(CT) scans. It offers a collection of annotated lung CT scans, where radiologists have marked and characterized lung nodules. This dataset helps researchers develop models that can accurately identify and classify lung nodules, supporting early lung cancer detection and diagnosis.
Camelyon16/17(breast cancer metastases detection)
The Camelyon16 and Camelyon17 datasets are significant in the domain of medical image analysis for breast cancer detection and metastasis identification. These datasets contain whole-slide pathology images of lymph nodes obtained from breast cancer patients. The aim is to detect metastases in these images, as identifying cancer spread to lymph nodes is crucial for determining cancer stage and treatment strategies. These datasets enable the training and evaluation of deep learning models to assist pathologists in detecting metastases accurately.
ISIC(skin lesion analysis)
The ISIC dataset focuses on skin lesion analysis and is essential for developing deep learning models for melanoma detection and skin lesion classification. It includes a wide range of images of skin lesions, along with expert annotations for lesion types and malignancy levels. This dataset supports the creation of models that aid dermatologists in early melanoma detection and skin cancer diagnosis.
MURA(musculoskeletal radiographs)
The MURA dataset provides musculoskeletal radiographs for training deep learning models to diagnose various bone and joint conditions. It includes X-ray images of different body parts and labels indicating whether the images are normal or abnormal. This dataset is valuable for creating models that can assist radiologists in identifying musculoskeletal disorders from X-ray images.
Challenges in Data Collection, Annotation, and Standardization: In medical imaging research, several challenges arise in data collection, annotation, and standardization. Data acquisition involves obtaining high-quality medical images with consistent imaging protocols, which can be hindered by variations in equipment, techniques, and patient conditions. Annotation, especially for pixel-level segmentation tasks, demands expert knowledge and is time-consuming. Standardizing annotations across experts can be challenging, leading to inter-annotator variability. Moreover, the scarcity of labeled data, especially in rare conditions, limits model training. Standardization of data formats, metadata, and ethical considerations for patient privacy further complicates research efforts.
Benchmarking results of deep learning models
ResNet, U-Net, DETR, and Inception models have shown promising results on various medical imaging datasets. For instance, ResNet's feature extraction capabilities contribute to high segmentation accuracy on brain tumor and lung nodule datasets. U-Net excels in skin lesion and lung nodule segmentation due to its specialized architecture. DETR's object detection capabilities lead to accurate detection of metastases in breast cancer datasets. Inception models showcase competitive segmentation accuracy across datasets due to their multi-scale feature extraction. Computational efficiency varies; U-Net's compact design allows quick training, while DETR's transformer architecture demands more time. Generalization capabilities depend on dataset diversity, with Inception models often demonstrating robustness across modalities.
Challenges in dataset acquisition and annotation
Dataset acquisition and annotation in medical imaging are challenging due to factors like limited access to sensitive patient data, requiring thorough ethical considerations. Expert annotations are critical for accurate model training, but the scarcity of skilled annotators can lead to delays. Additionally, bias can be introduced if annotations are subjective, potentially affecting model performance. Collecting diverse data across demographics and medical conditions is also problematic, impacting model generalization.
Potential solutions and best practices
To address these challenges, a collaborative approach involving healthcare professionals, researchers, and data scientists is crucial. Developing standardized protocols for data collection and annotation can reduce variability. Active learning can optimize annotation efforts by focusing on uncertain cases. Leveraging transfer learning from pre-trained models can mitigate data scarcity issues. Generating synthetic data through data augmentation techniques can enhance dataset diversity. Ensuring transparency in annotations and implementing bias detection mechanisms can mitigate bias risks.
Current trends and future directions
Recent developments like transfer learning, self-supervised learning, and multimodal fusion have elevated the capabilities of deep learning models in tumor detection and segmentation. Transfer learning, leveraging pre-trained models on large datasets, has improved model performance in scenarios with limited labeled medical data. Self-supervised learning techniques, where models learn from unannotated data, are gaining traction, reducing the reliance on manual annotations. Multimodal fusion, combining information from different imaging modalities, enhances accuracy by capturing complementary features.
Impact on clinical applications and research
These advancements have transformative implications for clinical applications and research. Transfer learning enables quicker model development, making AI-assisted diagnosis feasible in real-world clinical settings. Self-supervised learning addresses data scarcity issues, expanding the scope of medical image analysis to rare conditions. Multimodal fusion enhances diagnostic accuracy by integrating diverse information. These developments facilitate earlier and more accurate tumor detection, leading to better patient outcomes and improved treatment planning. Furthermore, they accelerate research by reducing the annotation burden and expanding the potential applications of AI in medical imaging.
Emerging trends and research direction
Future research in deep learning for tumor detection and segmentation is promising. Explainable AI methods aim to enhance model interpretability, enabling clinicians to understand model decisions and build trust. Federated learning ensures privacy by analyzing medical data locally and sharing aggregated insights, preserving patient confidentiality. Multi-task learning, where models simultaneously perform joint tasks like detection and segmentation, optimizes model efficiency and accuracy. Additionally, robustness to domain shifts, handling small datasets, and addressing bias remain active areas of investigation.
These advancements not only impact clinical practices by enabling accurate and efficient diagnoses but also drive research by expanding possibilities and reducing barriers. Future directions, including explainability and privacy-preserving techniques, will shape the evolving landscape of medical image analysis and foster continued innovation.
Discussion
Interdisciplinary collaboration among computer scientists, radiologists, pathologists, and clinicians is pivotal for propelling deep learning-based tumor analysis forward. This collaborative approach combines technical expertise, clinical insight, and data diversity to comprehensively address the complexities of tumor diagnosis. Computer scientists bring algorithmic prowess to create advanced models, while radiologists and pathologists provide specialized knowledge in interpreting medical images and histopathological slides. Clinicians contribute valuable contextual insights, ensuring that the developed solutions align with real-world clinical practices and patient care requirements. This multidisciplinary synergy enhances model accuracy, encourages ethical considerations, and promotes the development of interpretable models.
Despite its potential, deep learning-based tumor analysis faces challenges that interdisciplinary collaboration can help surmount. Rare tumor subtypes, often challenging to accurately diagnose, can benefit from diverse data sources and the combined expertise of pathologists and clinicians. Collaboration also aids in addressing model interpretability concerns by involving domain experts to develop models whose predictions can be understood and validated. Integrating these models into the clinical workflow requires input from all stakeholders to ensure seamless integration with existing practices and systems. Moreover, collaboration helps establish data quality standards, navigate regulatory requirements, and stay abreast of rapid advancements, positioning the field to make significant strides in cancer diagnosis and treatment.
Ethical Considerations
Ethical implications of using deep learning in tumor detection and medical imaging
The adoption of deep learning models for tumor detection and medical imaging comes with significant ethical considerations. Patient outcomes can be positively and negatively impacted by these models. While accurate models can lead to earlier and more precise diagnoses, improving treatment outcomes, erroneous predictions could result in delayed or incorrect treatments. The ethical responsibility lies in rigorously testing and validating models to ensure their reliability and safety before deploying them in clinical settings. Continuous monitoring, validation, and improvement are crucial to prevent potential harm to patients and maintain trust in the technology.
Patient privacy and data security concerns
The use of medical data for training and evaluating deep learning models raises privacy and security concerns. Anonymization techniques are essential to protect patient identities, but re-identification risks still exist. Collaborative efforts involving data sharing agreements between healthcare institutions and researchers can balance the need for data access with patient privacy. Compliance with data protection regulations such as HIPAA or GDPR is paramount to avoid legal repercussions. Ensuring secure data storage, transmission, and access control minimizes the risk of data breaches, safeguarding patients' sensitive medical information.
Bias and fairness in deep learning models for tumor analysis
Deep learning models can inherit biases from the data they are trained on, leading to disparities in predictions across different demographic groups or imaging modalities. Such biases could exacerbate healthcare disparities and lead to unequal access to accurate diagnosis and treatment. Ensuring fairness and inclusivity demands careful data collection, thorough analysis of potential biases, and ongoing monitoring of model performance across diverse populations. Developing representative and balanced datasets and using debiasing techniques are critical steps in reducing disparities and making AI-driven healthcare solutions equitable.
In conclusion, the ethical implications of using deep learning models in tumor detection and medical imaging require meticulous attention. Safeguarding patient outcomes, ensuring model safety, protecting privacy, addressing bias, and promoting fairness are central to the responsible development and deployment of AI-driven healthcare solutions. Collaborative efforts between medical professionals, computer scientists, ethicists, and regulatory bodies are essential to navigate these complex ethical considerations successfully.
Conclusion
The survey comprehensively explored the performance of ResNet, U-Net, DETR, and Inception variants in tumor detection and segmentation tasks. ResNet showcased its strength in feature learning, contributing to high accuracy particularly in intricate tumor patterns. U-Net excelled in segmenting small structures and demonstrated computational efficiency. DETR's transformer-based architecture proved effective in object detection tasks, while Inception models struck a balance between accuracy and efficiency. Each model exhibited distinct advantages, with trade-offs between accuracy, efficiency, and complexity.
Deep learning models have the potential to revolutionize clinical practice and research in tumor detection and segmentation. They offer accurate and efficient diagnoses, supporting clinicians in making informed decisions. These models contribute to improved patient care by enabling early detection, personalized treatment plans, and precise tumor localization. In research, deep learning accelerates the analysis of large datasets, fostering novel insights and facilitating the development of tailored therapies. The integration of AI-assisted tools enhances clinical workflow, reducing workload and ensuring better patient outcomes.
In conclusion, the survey underscores the significant strides made in applying deep learning models to tumor analysis. These models are not only advancing medical diagnostics but also reshaping the landscape of personalized patient care. As the field progresses, continued innovation is essential to tackle challenges, refine models, and expand their capabilities. Collaboration between clinicians, researchers, and data scientists will drive the adoption of these models in real-world healthcare settings. Ethical considerations, including data privacy and model interpretability, must remain at the forefront. The journey towards leveraging deep learning for tumor detection and segmentation is marked by promise, and sustained efforts will lead to transformative impacts in healthcare.
Source of Funding
None.
Conflict of Interest
None.
References
- X Kang, H Wang, J Guo, W Yu. Unsupervised deep learning method for color image recognition. J Comp Appl 2015. [Google Scholar]
- Y Lecun, Y Bengio, G Hinton. Deep learning. Nature 2015. [Google Scholar]
- JG Nam, S Park, EJ Hwang, JH Lee, KN Jin, KY Lim. Development and validation of deep learning-based automatic detection algorithm for malignant pulmonary nodules on chest radiographs. Radiology 2019. [Google Scholar]
- S Park, SM Lee, KH Lee, KH Jung, W Bae, J Choe. Deep learning-based detection system for multiclass lesions on chest radiographs: comparison with observer readings. Eur Radiol 2020. [Google Scholar]
- H Yoo, KH Kim, R Singh, SR Digumarthy, MK Kalra. Validation of a deep learning algorithm for the detection of malignant pulmonary nodules in chest radiographs. JAMA Netw Open 2020. [Google Scholar]
- Y Sim, MJ Chung, E Kotter, S Yune, M Kim, S Do. Deep convolutional neural network-based software improves radiologist detection of malignant lung nodules on chest radiographs. Radiology 2020. [Google Scholar]
- EJ Hwang, S Park, KN Jin, JI Kim, SY Choi, JH Lee. Development and validation of a deep learning-based automated detection algorithm for major thoracic diseases on chest radiographs. JAMA Netw Open 2019. [Google Scholar]
- R Manser, A Lethaby, LB Irving, C Stone, G Byrnes, MJ Abramson. Screening for lung cancer. Cochrane Database Syst Rev 2013. [Google Scholar]
- L Berlin. Radiologic errors, past, present and future. Diagnosis (Berl) 2014. [Google Scholar]
- Y Lecun, B Boser, JS Denker, D Henderson, RE Howard, W Hubbard. Backpropagation applied to handwritten zip code recognition. Neural computation. Neural Comput 1989. [Google Scholar]
- Z Liu, H Mao, CY Wu, C Feichtenhofer, T Darrell, S Xie. A convnet for the 2020s. . [Google Scholar] [Crossref]
- P Lakhani, B Sundaram. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 2017. [Google Scholar]
- JK Min, MS Kwak, JM Cha. Overview of deep learning in gastrointestinal endoscopy. Gut Liver 2019. [Google Scholar]
- T Würfl, FC Ghesu, V Christlein, A Maier. Deep learning computed tomography. 2016. [Google Scholar] [Crossref]
- MM Lell, M Kachelrieß. Recent and upcoming technological developments in computed tomography: high speed, low dose, deep learning, multienergy. Investigative radiology. Invest Radiol 2020. [Google Scholar]
- A Hamidinekoo, E Denton, A Rampun, K Honnor, R Zwiggelaar. Deep learning in mammography and breast histology, an overview and future trends. Med Image Anal 2018. [Google Scholar]
- F Liu, S Ge, Y Zou, X Wu. Competence-based Multimodal Curriculum Learning for Medical Report Generation. 2023. [Google Scholar] [Crossref]
- AS Lundervold, A Lundervold. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 2019. [Google Scholar]
- Z Akkus, A Galimzianova, A Hoogi, DL Rubin, BJ Erickson. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017. [Google Scholar]
- AJ Reader, G Corda, A Mehranian, CDa Costa-Luis, S Ellis, J Schnabel. Deep learning for PET image reconstruction. es 2020. [Google Scholar]
- K He, X Zhang, S Ren, J Sun. Deep residual learning for image recognition. InProceedings of the IEEE conference on computer vision and pattern recognition 2016. [Google Scholar] [Crossref]
- J Li, F Fang, K Mei, G Zhang. Multi-scale residual network for image super-resolution. 2018. [Google Scholar]
- A Murad, GR Kwon, JY Pyun. P2-402: Brain age prediction from minimally preprocessed mri scans using 3d deep residual neural networks. Alzheimers Dement 2018. [Google Scholar] [Crossref]
- A Nibali, Z He, D Wollersheim. Pulmonary nodule classification with deep residual networks. Int J Comput Assist Radiol Surg 2017. [Google Scholar]
- A Maier, C Syben, T Lasser, C Riess. A gentle introduction to deep learning in medical image processing. Z Med Phys 2019. [Google Scholar]
- R Karthik, R Menaka, M Hariharan. Learning distinctive filters for COVID-19 detection from chest X-ray using shuffled residual CNN. Appl Soft Comput 2021. [Google Scholar]
- Y Lu, X Qin, H Fan, T Lai, Z Li. WBC-Net: A white blood cell segmentation network based on UNet++ and ResNet. Applied Soft Computing 2021. [Google Scholar]
- A Nazir, MN Cheema, B Sheng, H Li, P Li, Po Yang. OFF-eNET: An optimally fused fully end-to-end network for automatic dense volumetric 3D intracranial blood vessels segmentation. IEEE Transactions on Image Processing 2020. [Google Scholar] [Crossref]
- J Arevalo, FA González, RR Pollán, JL Oliveira, MAG Lopez. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed 2016. [Google Scholar]
- T Zhang, S Zheng, J Cheng, Xi Jia, J Bartlett, X Cheng. Structure and Intensity Unbiased Translation for 2D Medical Image Segmentation. IEEE Trans Pattern Anal Mach Intell 2024. [Google Scholar] [Crossref]
- Ö Çiçek, A Abdulkadir, SS Lienkamp, T Brox, O Ronneberger. 3D U-Net: learning dense volumetric segmentation from sparse annotation. 19th International Conference 2016. [Google Scholar] [Crossref]
- LH Schwartz, S Litière, ED Vries, R Ford, S Gwyther, S Mandrekar. RECIST 1.1-Update and clarification: From the RECIST committee. Eur J Cancer 2016. [Google Scholar] [Crossref]
- Q Dou, H Chen, Y Jin, H Lin, J Qin, PA Heng. Automated pulmonary nodule detection via 3d convnets with online sample filtering and hybrid-loss residual learning. InMedical Image Computing and Computer Assisted Intervention 2017. [Google Scholar] [Crossref]
- Z Liu, Y Lin, Y Cao, H Hu, Y Wei, Z Zhang. Swin transformer: Hierarchical vision transformer using shifted windows. InProceedings of the IEEE/CVF international conference on computer vision 2021 2021. [Google Scholar] [Crossref]
- N Carion, F Massa, G Synnaeve, N Usunier, A Kirillov, S Zagoruyko. End-to-End Object Detection with Transformers. 2020. [Google Scholar] [Crossref]
- C Szegedy, V Vanhoucke, S Ioffe, J Shlens, Z Wojna. Rethinking the inception architecture for computer vision. InProceedings of the IEEE conference on computer vision and pattern recognition 2015. [Google Scholar] [Crossref]
- L Wang, Y Xiong, Z Wang, Y Qiao, D Lin, X Tang. Temporal Segment Networks: Towards Good Practices for Deep Action Recognition. 2016. [Google Scholar] [Crossref]
- Introduction
- Deep learning models
- Overview of tumor detection and segmentation
- Deep learning fundamentals
- ResNet and variants
- Overview of ResNet architecture
- Applications of ResNet in tumor detection and segmentation
- Variants and modifications of ResNet for tumor analysis
- U-Net and variants
- U-Net architecture
- U-Net for limited labeled data
- Applications of U-Net in tumor detection and segmentation
- Adaptations and enhancements of U-Net
- DETR(Detection Transformer)
- DETR architecture and object detection
- Advantages and limitations of using DETR
- Adaptations and extensions of DETR for tumor detection and segmentation
- Inception and variants
- Inception module and computational complexity
- Performance of inception models
- Variants and customizations of inception models for medical imaging
- Benefits of inception variants in tumor analysis
- Comparative analysis
- ResNet
- U-Net
- DETR
- Inception models
- Trade-offs and considerations
- Datasets and benchmarking
- BraTS(brain tumor segmentation)
- LIDC-IDRI(lung nodule detection)
- Camelyon16/17(breast cancer metastases detection)
- ISIC(skin lesion analysis)
- MURA(musculoskeletal radiographs)
- Benchmarking results of deep learning models
- Challenges in dataset acquisition and annotation
- Potential solutions and best practices
- Current trends and future directions
- Impact on clinical applications and research
- Emerging trends and research direction
- Discussion
- Ethical Considerations
- Ethical implications of using deep learning in tumor detection and medical imaging
- Patient privacy and data security concerns
- Bias and fairness in deep learning models for tumor analysis
- Conclusion
- Source of Funding
- Conflict of Interest
